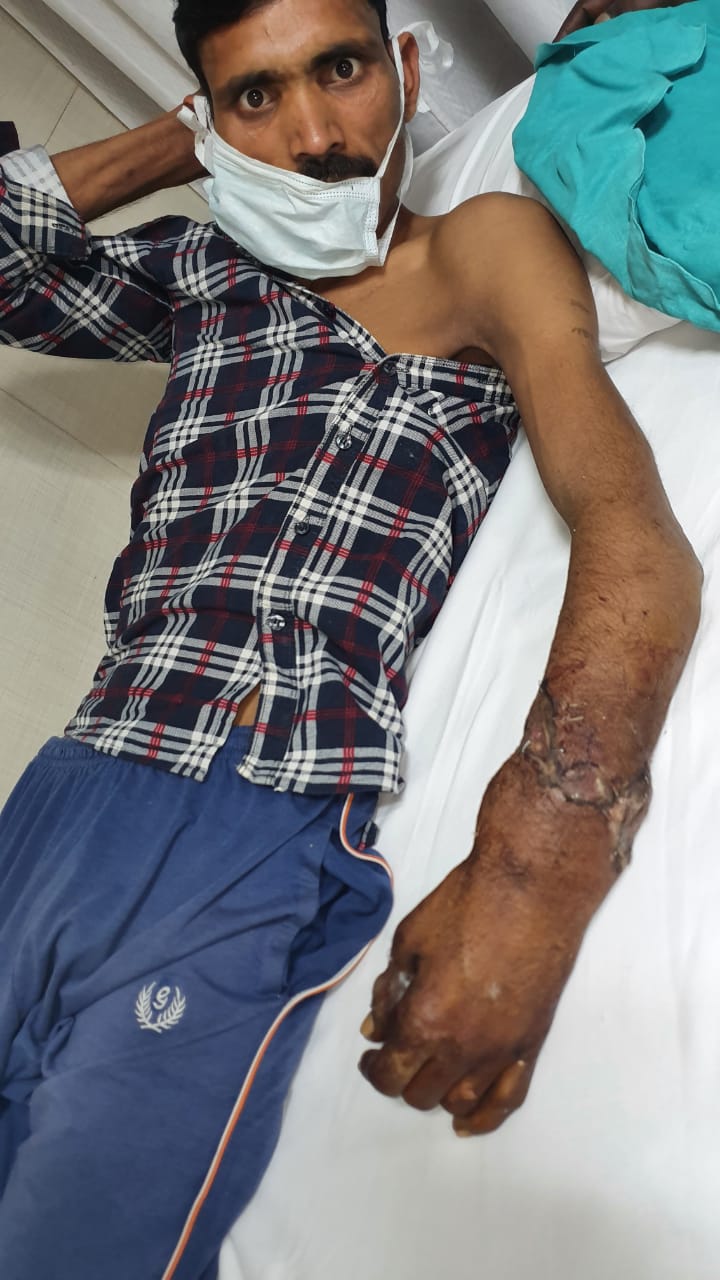
New Delhi – A 36 years old Inderpal while working in a factory in Prahaladpur Industrial Area, Badli, New Delhi , suddenly a heavy object from the machine fell on his hand and his left forearm got crushed and amputated. He suffered from severe pain , loss of blood and went into the shock. Immediately after the injury he was taken to the hospital for treatment that saved his amputated hand.
“Though the patient reached us well within the golden time period (usually 3 to 4 hours over ice in forearm level injury), it was challenging as the forearm at the time of amputation got badly pulled and crushed. This led to multiple level injuries to the various structures (bone, muscles, nerves and vessels), necessitating a holistic approach requiring extensive shortening and graft usage from different parts of the body.” said Dr. Anubhav Gupta, Senior Consultant, Department of Plastic and Reconstructive Surgery, Sir Ganga Ram Hospital
“The latest surgical techniques it is now possible to re-join even the worst amputated limbs if they are brought in time and well preserved in ice. It is important to understand that the amputated part should not be in direct contact of ice. It should be kept in a clean polythene over ice.” Dr. Anubhav Gupta further explained
Due to the severity of the injury, it took us over 6 hours to perform the implantation of the forearm using micro-vascular technique. By 12 midnight the patient was wheeled out form the operation theatre.
At present the patient’s hand has been successfully salvaged and he hopes to have a good hand functional recovery.